Recent stages of the Mapping Cancer Markers (MCM) project have illuminated the protein-protein interactions and biological pathways involved in lung cancer, and have also suggested surprising results about its biomarkers. Once this current stage is complete, MCM will transition to analyzing ovarian cancer. Thanks to your help, we are making discoveries and helping the international research community. Dr. Jurisica, in particular, is one of the most frequently cited researchers worldwide.
In our previous update, we announced a second, targeted stage of lung cancer signature discovery. We have since moved to a new, third stage in lung cancer analysis: targeting high-scoring, uncorrelated biomarkers. These different stages are all part of an overall effort to understand lung cancer signatures. The first stage surveyed possible lung cancer signatures drawn from the complete set of biomarkers in our lung cancer dataset. The statistics gathered in this first stage were used to narrow the list of biomarkers to explore in subsequent stages. The second and third stages explore lung cancer signatures drawn from small sets of high-performing signatures, chosen by two different methods. In the second stage, we focused on a 1% subset of biomarkers, selected by the frequency with which each appeared in high-scoring signatures from the initial stage. In the third stage, we selected a different subset of biomarkers that are both high-scoring and largely uncorrelated to one another.
Correlation is a measure of information shared between two data sources. Two biomarkers are correlated if they exhibit similar patterns in the cancer dataset. For example, two correlated genes might show high activity in one set of tumour samples, low activity in a second set, and average activity in a third. Including two highly-correlated biomarkers in the same signature can reduce the quality of the signature, because they would be contributing redundant information to the signature. For a fixed-size signature, a redundant biomarker would potentially displace another biomarker that has different information content.
As an analogy, consider the information contained in a small library of textbooks. Say there are three books, A, B, and C. If A and B are two copies of the same textbook, one of them is redundant. Removing B from the library would not change the information contained in the library, and replacing B with a different textbook (D), would increase the information in the library. If A and B were similar, but not identical books (e.g., two books on introduction to molecular biology written by different authors), there would still be some overlap in the texts, and a possible advantage to replacing B with D.
Signature performance
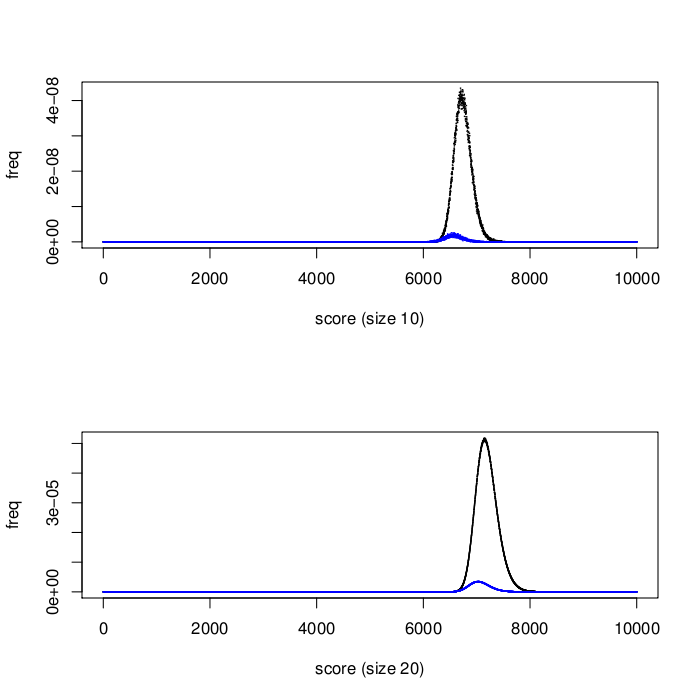
Because the target biomarkers in this third stage were selected to be minimally inter-correlated, every signature should be free of redundant information. We therefore hypothesized that signatures in the third stage would perform better on average than those in the second stage. Figure 1 shows the surprising results: second stage signatures (potentially containing correlated biomarkers) outperformed those from the third stage. We are analysing these results further, to determine the main reasons for the performance difference.
Size effects on biomarker rank in top signatures
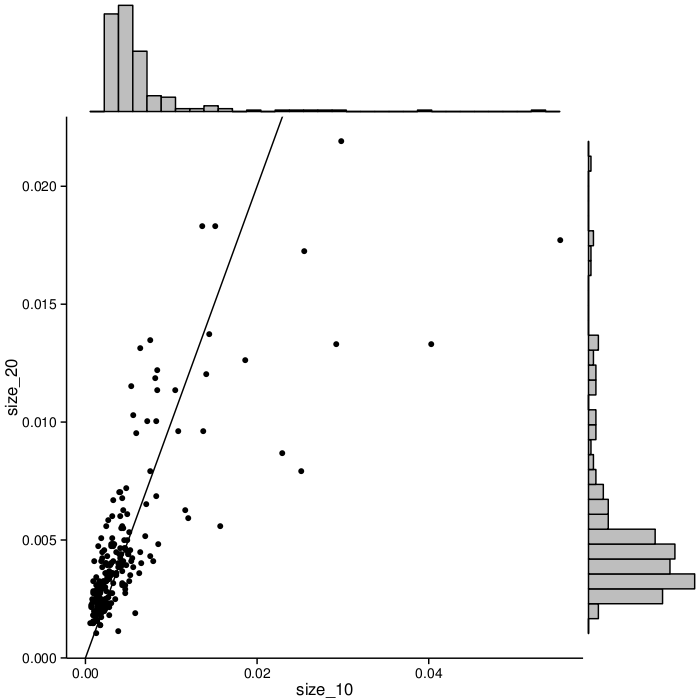
Larger signatures (i.e., signatures containing more biomarkers) incorporate more information and can potentially offer better accuracy, but are more complex and expensive to implement in the clinic. All three stages of MCM thus far have explored lung cancer signatures of multiple sizes. For each signature size we considered, the target biomarker subsets for the second stage were chosen separately, based on statistics from the first stage. The set of biomarkers selected for the third is fixed across all signature sizes. This fixed set allows us to compare the effects of signature size on each biomarker's frequency in high-scoring signatures. Figure 2 shows the frequency change when moving from 10 biomarkers per signature to 20. Each dot in the graph represents a biomarker. The X axis represents the frequency with which biomarkers appear in size_10 signatures. The Y axis indicates frequency in size_20 signatures. Note that the biomarkers change in rank but are generally correlated. Size_10 signatures show greater biomarker frequency spread: some have relatively high frequency, and many are low-frequency. The biomarker frequencies in larger (size_20) signatures are more even.
Biomarker pairs as protein interactions?
We applied and extended the analysis of biomarker pairs described in the August 2015 update to early results from third stage data, looking specifically for pairs of biomarkers in both the second and third stages that appear surprisingly frequently in the highest-scoring lung cancer signatures. When two genes or proteins appear in signatures together with greater frequency than expected randomly, we predict a stronger cancer-related connection (interaction).
We searched for any known connections (interactions) in The Integrated Interactions Database (IID), a database of known and predicted protein-protein interactions created by our lab [1]. We found several interactions in IID that mirror these cancer interactions, but the overlap was not statistically significant.
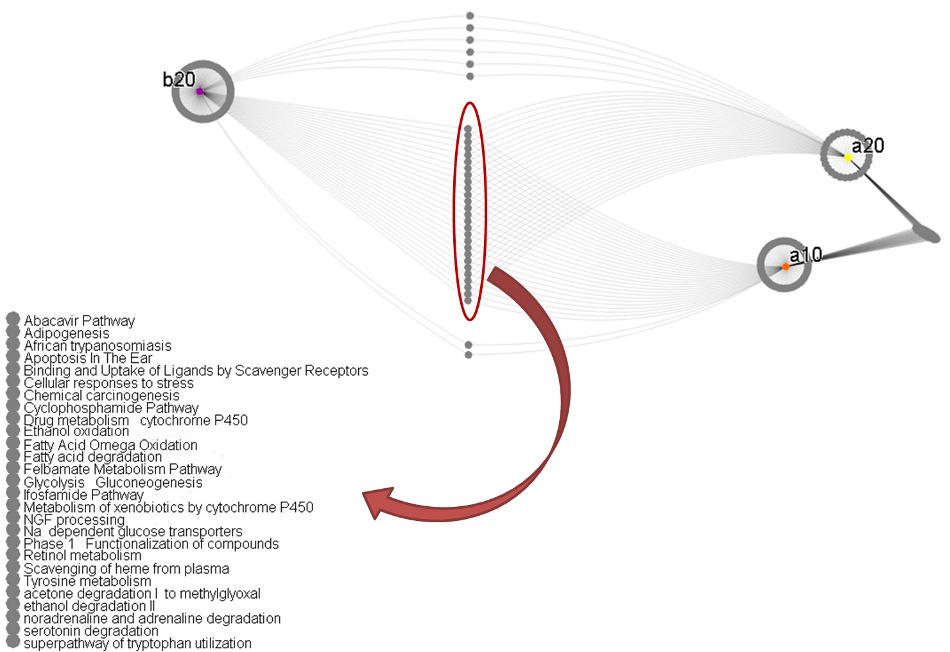
Pathway enrichment in second and third stage targets
We also took the genes selected for the second and third stages, and searched for them in a database of biological pathways. See Figure 3. We discovered our lists of genes were enriched (present in statistically significant numbers; p ≤ 0.01) in several pathways. See Table 1.
Although our analysis is ongoing, we can see that two of the identified pathways are components of Mevalonate metabolism. Mevalonate pathways are already targets for many drugs such as statins and have been implicated as targets for treatment in lung cancer [2, 3]. Some of the downstream analysis will focus on how the signatures discovered by World Community Grid processing will ultimately connect to pathways and other research. We have used Mevalonate as an example, but there are many more that can be examined to assess the viability of our best signatures.
Table 1. List of biological pathways enriched with MCM's "discovered-pair" genes. P-values < 0.01 indicate statistical significance.
| Pathway Name | p-value |
|---|---|
| Mevalonate from acetyl CoA step 2 3 | 0.003236 |
| Biotinidase Deficiency metabolite pathway | 0.004845 |
| Biotin Metabolism | 0.004845 |
| Biotinidase Deficiency | 0.004845 |
| Multiple carboxylase deficiency neonatal or early onset form | 0.004845 |
| Mevalonate biosynthesis | 0.004845 |
| Synthesis of Ketone Bodies | 0.006449 |
| Ketone Body Metabolism | 0.008048 |
| Succinyl CoA 3 ketoacid CoA transferase deficiency | 0.008048 |
| Synthesis and Degradation of Ketone Bodies | 0.01 |
| Fatty acid triacylglycerol and ketone body metabolism | 0.008892 |
| Vitamin H biotin metabolism | 0.009643 |
| Dermatan sulfate degradation metazoa | 0.009643 |
Transition from lung cancer to ovarian cancer analysis
The third stage is nearly complete, and will be the final piece of MCM lung cancer analysis on World Community Grid before we switch to ovarian cancer.
Ovarian cancer is a gynecologic malignancy that ranks 8th for incidence and 5th for death rate among all women's cancers. The American National Cancer Institute's Surveillance, Epidemiology, and End Results (SEER) program estimated 22,240 new cases and 14,030 deaths from ovarian cancer in 2013. Patients are usually diagnosed at an advanced stage (61% present metastasized cancer) and have poor prognosis (27.3 months for metastasized stage (SEER)).
Ovarian cancer was chosen as our next dataset because of long experience with this disease in our own lab, and in those of collaborators. We look forward to using MCM to glean new insights into ovarian cancer.
We expect the transition to ovarian cancer research to begin in early 2016, and do not anticipate any interruption in the flow of work units.
Thank you to World Community Grid members
We wish to thank World Community Grid members for their continued support and interest for this and other projects. Without you, this work would not be possible.
References
-
1. Kotlyar M, Pastrello C, Sheahan N, Jurisica I. Integrated interactions database: tissue-specific view of the human and model organism interactomes. Nucleic Acids Res. 2015 Oct 29
2. Hwa Young Lee, In Kyoung Kim, Hye In Lee, Hye Sun Kang, Chan Kwon Park, Jick Hwan Ha, Seung Joon Kim, Sang Haak Lee. Mevalonate pathway inhibitors as chemopreventive agents on lung cancer cell lines: p53 might be a potent regulator. [abstract]. In: Proceedings of the Eleventh Annual AACR International Conference on Frontiers in Cancer Prevention Research; 2012 Oct 16-19; Anaheim, CA. Philadelphia (PA): AACR; Cancer Prev Res 2012;5(11 Suppl):Abstract nr A48.
3. Yano K. Lipid metabolic pathways as lung cancer therapeutic targets: a computational study. Int J Mol Med. 2012 Apr;29(4):519-29. doi: 10.3892/ijmm.2011.876. Epub 2011 Dec 30.
In several papers we have used strategies described above and protein interaction networks to identify better prognostic markers and new treatment options:
- Singh M, Garg N, Venugopal C, Hallett RM, Tokar T, McFarlane N, Arpin C, Page B, Haftchenary S, Todic A, Rosa DA, Lai P, Gómez-Biagi R, Ali AM, Lewis A, Geletu M, Mahendram S, Bakhshinyan D, Manoranjan B, Vora P, Qazi M, Murty NK, Hassell JA, Jurisica I, Gunning P, Singh SK. STAT3 pathway regulates lung-derived brain metastasis initiating cell capacity through miR-21 activation. Oncotarget (accepted June 30, 2015, ONC-2014-02546)
- Navab R, Strumpf D, To C, Pasko E, Kim KS, Park CJ, Hai J, Liu J, Jonkman J, Barczyk M, Bandarchi B, Wang YH, Venkat K, Ibrahimov E, Pham NA, Ng C, Radulovich N, Zhu CQ, Pintilie M, Wang D, Lu A, Jurisica I, Walker GC, Gullberg D, Tsao MS. Integrin a11b1 regulates cancer stromal stiffness and promotes tumorigenecity in non-small cell lung cancer, Oncogene, 2015. In press.
- Agostini M, Zangrando A, Pastrello C, D'Angelo E, Romano G, Giovannoni R, Giordan M, Maretto I, Bedin C, Zanon C, Digito M, Esposito G, Mescoli C, Lavitrano M, Rizzolio F, Jurisica I, Giordano A, Pucciarelli S, Nitti D. A functional biological network centered on XRCC3: a new possible marker of chemoradiotherapy resistance in rectal cancer patients, Cancer Biol Ther, 16(8):1160-71, 2015.
- Agostini M, Janssen KP, Kim LJ, D'Angelo E, Pizzini S, Zangrando A, Zanon C, Pastrello C, Maretto I, Digito M, Bedin C, Jurisica I, Rizzolio F, Giordano A, Bortoluzzi S, Nitti D, Pucciarelli S. An integrative approach for the identification of prognostic and predictive biomarkers in rectal cancer. Oncotarget. 2015. Sep 2.
- Stewart, E.L., Mascaux, C., Pham, N-A, Sakashita, S., Sykes, J., Kim, L., Yanagawa, N., Allo, G., Ishizawa, K., Wang, D., Zhu, C.Q., Li, M., Ng, C., Liu, N., Pintilie, M., Martin, P., John, T., Jurisica, I., Leighl, N.B., Neel, B.G., Waddell, T.K., Shepherd, F.A., Liu, G., Tsao, M-S. Clinical Utility of Patient Derived Xenografts to Determine Biomarkers of Prognosis and Map Resistance Pathways in EGFR-Mutant Lung Adenocarcinoma, J Clin Oncol, 33(22):2472-80, 2015.
- Camargo, J. F., Resende, M., Zamel, R., Klement, W., Bhimji, A., Huibner, S., Kumar, D., Humar, A., Jurisica, I., Keshavjee, S., Kaul, R., Husain, S. Potential role of CC chemokine receptor 6 (CCR6) in prediction of late-onset CMV infection following solid organ transplant. Clinical Transplantation, 2015. In press. doi: 10.1111/ctr.12531
- Fortney, K., Griesman, G., Kotlyar, M., Pastrello, C., Angeli, M., Tsao, M.S., Jurisica, I. Prioritizing therapeutics for lung cancer: An integrative meta-analysis of cancer gene signatures and chemogenomic data, PLoS Comp Biol, 11(3): e1004068, 2015.
- Benleulmi-Chaachoua, A., Chen, L., Sokolina, K., Wong, V., Jurisica, I., Emerit, M.B., Darmon, M., Espin, A., Stagljar, I., Tafelmeyer, P., Zamponi, G.W., Delagrange, P., Maurice, P., Jockers, R. Protein interactome mining defines melatonin MT1 receptors as integral component of presynaptic protein complexes of neurons, Journal of Pineal Research, In press
Media Coverage
Also, for the second year in a row, Dr. Jurisica has been included in Thomson Reuters highly cited researcher list; Out of 108 in computer science and 3,125 world-wide in 21 fields of science.
Related Articles
- One step closer to identifying lung cancer biomarkers
- Mapping Cancer Markers Begins Analyzing Lung and Ovarian Cancer Data
- Mapping Cancer Markers Team Analyzes Lung Cancer Data
- New Facility and Expanded Plans for the Mapping Cancer Markers Research Team
- Sarcoma Dataset Coming Soon to Mapping Cancer Markers Project